Highly Scalable System for DNA Analysis
Description
The project is a pyrosequencing system that enables high-resolution detection and analysis of biomaterial for genetic mutations. As a result of cooperation with Altoros, the customer got:
- A scalable system that can process 10,000+ DNA samples at a time (10x more than their legacy framework).
- Reduced time spent on analysis: shortened from hours to minutes.
- A reference architecture for a reporting solution based on open-source technologies, saving thousands of dollars on costly BI licenses.
The customer
QIAGEN is a global provider of sample and assay technologies for molecular diagnostics, applied testing, and academic/pharmaceutical research. Its solutions help to transform biological material into valuable molecular insights. Headquartered in the Netherlands, the company operates 35 offices worldwide with 4,000+ employees, serving 500,000+ customers. Examples of expertise:
- Next-generation sequencing (NGS), a method that makes decoding genetic information much easier, faster, and cheaper than conventional methods.
- Pyrosequencing, a high-resolution detection technology that enables real-time analysis of biomaterial for possible genetic mutations.
The need
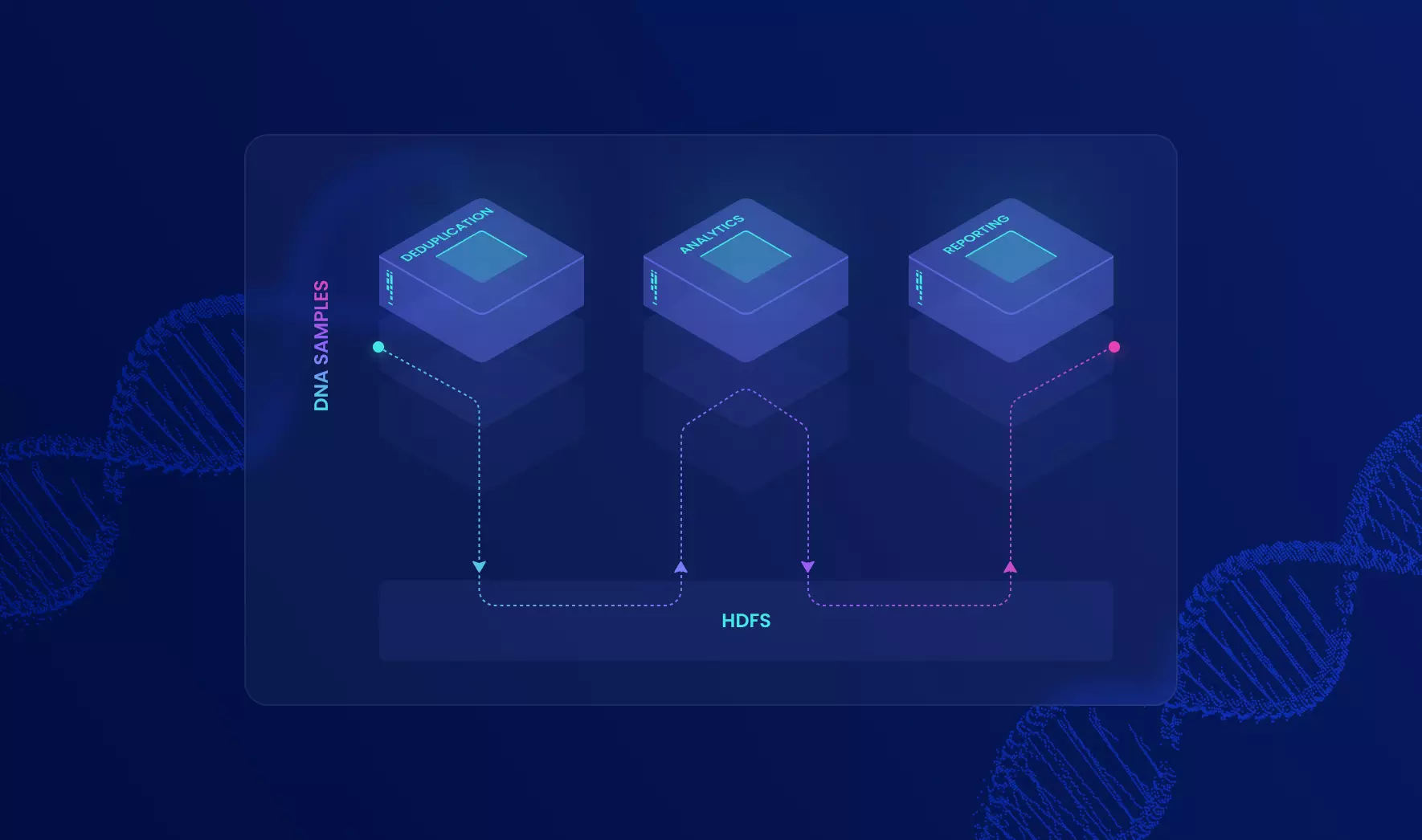
The customer turned to Altoros to improve its biotechnology system that analyzes DNA samples for mutations in the early stages. The legacy tool de-duplicated only 1,000 samples maximum — due to memory and CPU limitations — and it still took hours (or even days) to process the pipeline. The goal was to fix performance bottlenecks and enable linear scalability for processing 10,000+ biosamples at a time.
The challenges
Since the existing system was already operating on the superior hardware, vertical scaling was no longer an option. The team was also challenged to identify the legacy code parts that would allow for parallel processing of DNA samples with Hadoop. Finally, the system should have been seamlessly migrated to production.
The solution
Working as an extension of the customer’s team at their U.S. offices, our Java engineers assisted in installing and configuring Cloudera CDH 5.2 for distributed data storage and computation. Cluster monitoring and profiling were enabled with Cloudera Manager. After that, our developers have created a mini-framework — based on MapReduce jobs — with custom partitioners that enabled efficient distribution of data between parallel tasks. The team has also built a converter that transforms binary variant files (samples) into the Hadoop sequence format — required by the HDFS file system. In addition, we designed a reference architecture for an improved reporting solution integrated with Apache Spark. Our experts suggested using Spark SQL to preserve the reporting module's existing structure (SQL-based) and change data sources if needed easily.
The outcome
Altoros has delivered a highly scalable analytical system for de-duplication of genome samples - as a part of the customer’s analytical platform. Thousands of hospitals and laboratories worldwide use the system to detect DNA mutations, saving thousands of lives. The analysis takes minutes now, not hours; it allows for processing 10x more genome samples compared to the performance of the legacy system.
Altoros’s engineers have also proposed a reference architecture for updating a reporting solution. Inspired by our recommendations, the customer went on improving the system with open-source data analytics technologies, which will eventually allow for saving thousands of dollars on expensive Oracle BI licenses.
Server platform
Linux
Programming languages
Java, Perl
Technologies
Apache Hadoop (Cloudera CDH 5.2.1), MapReduce, Apache Spark (Spark SQL), bash
Databases
HDFS
Seeking a solution like this?
Contact us and get a quote within 24 hours